SCIENCE
What we do
The human genome project found that 1% of our genome is translated into protein and yet 80% is transcribed into RNA. It is recognized that RNA plays key roles in biology by not only encoding proteins, but also by various non-coding functions such as epigenetic regulation and cellular signaling. Therefore, the number of RNA drug targets is >100-fold greater than that of proteins, and the ability to modulate RNA function by small molecules can broadly impact drug discovery by both the development of precision medicines and the elucidation of fundamental disease biology.


The Disney group’s central focus is the development of small molecule precision medicine targeting RNA. We use the compounds that emerge from these studies to elucidate the rules of molecular recognition between small molecules and RNA. Furthermore, by applying these compounds to diseased and healthy cells, we can study the roles of RNA in biological processes to better understand this important class of biomolecule. Our studies have developed compounds that target RNA in a broad range of diseases with no cures or poor prognosis, ranging from cancers to Alzheimer’s, and from amyotrophic lateral sclerosis (ALS) to muscular dystrophy.
Furthermore, our group recognized that protein-targeted medicines can bind RNA and affect RNA-mediated pathways, underscoring that RNA should be considered as an on- or off-target in drug development. The work of our group has broadly influenced the pharmaceutical industry as they adopt many of our methods and platforms. Our group members go on to leadership roles in academia and industry and use science as a vehicle to produce our greatest impact to the world. Below we list some areas of focus in our lab, but these topics evolve periodically, and the most recent work can be found at our recent publications.
Sequence-based drug design
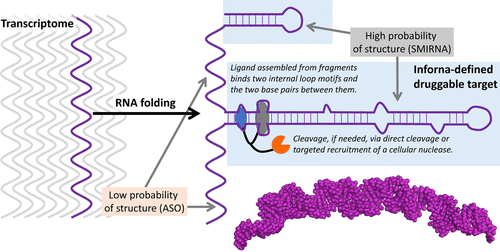
Since 2005, we have developed an approach to identify folded RNAs and to quickly design small molecules that selectively target these structures. These studies were powered by our group’s work on studying interactions between small molecules and RNA targets by using evolutionary principles, profiling diverse collections of small molecules for binding to libraries of RNA targets. Not only could we identify these binding interactions, but also we have developed computational methods to score their affinity in an unbiased manner. These data sets are computationally mined across the human transcriptome (the RNAs produced from a genome) to identify an RNA fold and a small molecule that can target it.
-
Sequence-based design of bioactive small molecules that target precursor microRNAs | Nature Chemical Biology
-
A Small Molecule Microarray Platform To Select RNA Internal Loop−Ligand Interactions | ACS Chemical Biology
-
Inforna 2.0: A Platform for the Sequence-Based Design of Small Molecules Targeting Structured RNAs | ACS Chemical Biology
-
DNA-encoded library versus RNA-encoded library selection enables design of an oncogenic noncoding RNA inhibitor | PNAS
Design of small molecules targeting cancer
One focus of our group’s efforts has been to identify small molecules that can bind to and disable RNAs that play important roles in cancer. Since many cancers lack a precision medicine that specifically targets a cancer-causing gene product, our group’s goal is to design and deliver different compounds that target RNAs that cause cancer. This approach can enable tailoring a precision medicine or cocktail to treat the cancer based on a patient’s genome. Many of these compounds are evaluated not only in patient-derived cancer cells but also in pre-clinical models to push these medicines closer to human clinical trials.
-
Programming inactive RNA-binding small molecules into bioactive degraders | Nature
-
Design of a small molecule against an oncogenic noncoding RNA | PNAS
-
Transcriptome-Wide Mapping of Small-Molecule RNA-Binding Sites in Cells Informs an Isoform-Specific Degrader of QSOX1 mRNA | Journal of the American Chemical Society
-
DNA-encoded library versus RNA-encoded library selection enables design of an oncogenic noncoding RNA inhibitor | PNAS
Design of small molecules targeting mRNA encoding “undruggable” proteins

Our group has also focused on targeting proteins that are considered “undruggable” by disabling their mRNAs from being translated or spliced to form toxic proteins. We have applied these approaches broadly and developed new methods to affect translation and to control pre-mRNA splicing outcome. For example, in Parkison’s disease, overexpression of the protein, alpha synuclein, leads to neuron death. However, this protein lacks a targetable structure by small molecules. The mRNA that encodes for alpha synuclein protein, however, does fold into a structure near its start codon. A compound that binds to this structure selectively reduces the levels of alpha synuclein protein and provides cytoprotective effects against alpha synuclein-induced apoptosis. In another example of Alzheimer’s and dementia, Tau protein can be toxic when it is in an aggregation-prone 4R form. The pre-mRNA that encodes for Tau protein has a structure that can be modulated to affect the splicing of pre-mRNA and therefore to reduce the production of toxic 4R tau. Small molecules designed to target this pre-mRNA reduce 4R Tau in both cellular and pre-clinical models. We have advanced these approaches to a variety of other targets and significantly expanded the “druggability” of the human genome.
Augmenting the function of RNA binders
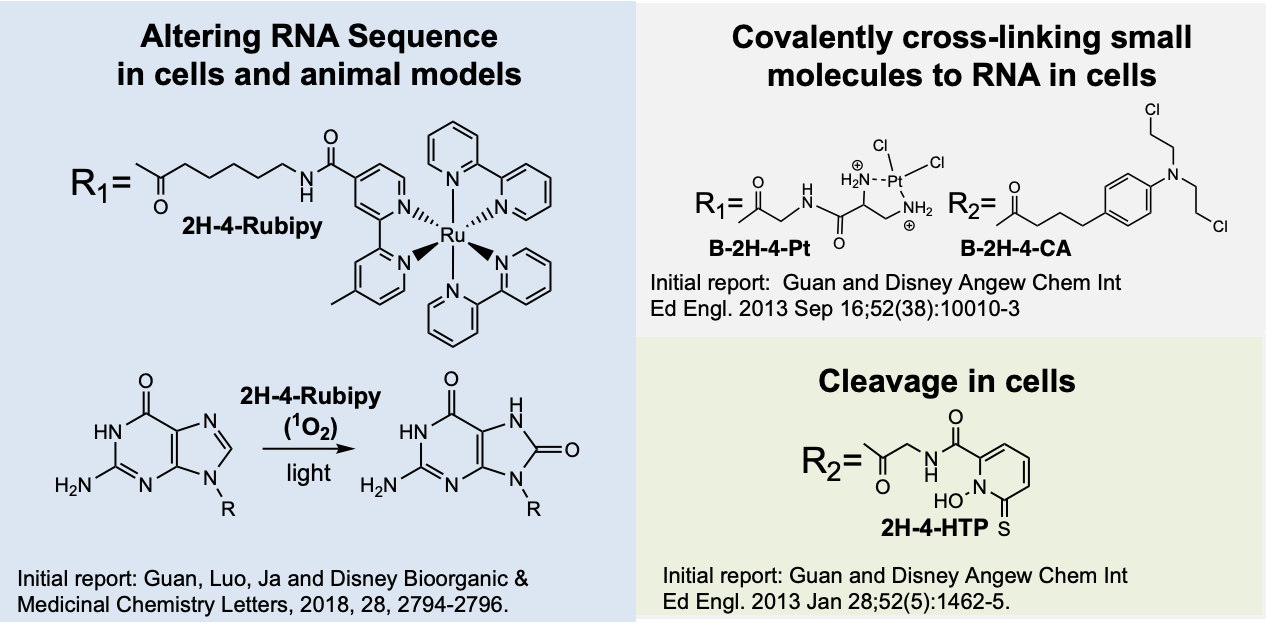

Over the past decade, we have developed various approaches to augment the function of RNA binders. For example, we have garnered perhaps the most attention by recruitment of enzymes to affect RNA lifetime via a ribonuclease targeting chimera (RiboTAC) approach. We have also developed direct degrader technologies where we append RNA binders with compounds that cause oxidative cleavage of RNA in cells and pre-clinical models without enzyme recruitment. Additionally, we have developed the first compounds that undergo cross-linking to RNA in cells to affect biological function, as well as methods to use small molecules to alter RNA sequence both in vitro and in vivo. Much will be continued in this area and many in the industry have adopted these approaches for target validation of RNA-small molecule interactions and to cleave RNA targets for developing new medicines.
RiboTACs:
-
Programming inactive RNA-binding small molecules into bioactive degraders | Nature
-
Small Molecule Targeted Recruitment of a Nuclease to RNA | Journal of the American Chemical Society
-
Small-molecule targeted recruitment of a nuclease to cleave an oncogenic RNA in a mouse model of metastatic cancer | PNAS
Direct cleavers of RNA:
-
Small-Molecule-Mediated Cleavage of RNA in Living Cells | Angewandte Chemie
-
Precise small-molecule cleavage of an r(CUG) repeat expansion in a myotonic dystrophy mouse model | PNAS
-
Precise Small Molecule Degradation of a Noncoding RNA Identifies Cellular Binding Sites and Modulates an Oncogenic Phenotype | ACS Chemical Biology
Covalent bond formation, cross linkers: